DPE: Difference between revisions
(Created page with "Data processing enegine serves for a processing of data acquired with the Timepix detectors (TPX, TPX2, TPX3). The following list includes the main processing parts of the DPE: * '''Pre-processing''': clustering of data with application of calibration and corrections, calculation of cluster variables/parameters, creation of histograms * '''Processing''': particle recognition * '''Post-processing''': calculation of physical products - fluxes, dose rates etc. This docume...") |
No edit summary |
||
| Line 22: | Line 22: | ||
The program especially in the case of graphic export needs several hundreds of MB on RAM (app 500 MB but it is based on the size of exported histograms). If the multiprocessing is used then the RAM usage can be up several GB and all threads of the CPU are used. | The program especially in the case of graphic export needs several hundreds of MB on RAM (app 500 MB but it is based on the size of exported histograms). If the multiprocessing is used then the RAM usage can be up several GB and all threads of the CPU are used. | ||
=== Run the program on linux: === | |||
<pre class="sh"> chmod +x dpe.sh | <pre class="sh"> chmod +x dpe.sh | ||
./dpe.sh PARAMETERS_FILE_PATH/PARATERS_FILE_NAME</pre> | ./dpe.sh PARAMETERS_FILE_PATH/PARATERS_FILE_NAME</pre> | ||
It is possible that also clusterer has to be allowed as an executable: <code>chmod +x clusterer</code>. The current build for linux was made with gcc version 7.5.0 on Ubuntu 18. | It is possible that also clusterer has to be allowed as an executable: <code>chmod +x clusterer</code>. The current build for linux was made with gcc version 7.5.0 on Ubuntu 18. | ||
=== Run the program on windows: === | |||
<pre class="ps"> DPE.exe PARAMETERS_FILE_PATH\PARATERS_FILE_NAME</pre> | <pre class="ps"> DPE.exe PARAMETERS_FILE_PATH\PARATERS_FILE_NAME</pre> | ||
If your path or name of the parameters includes white spaces enclose the in quotes: "PARAMETERS_FILE_PATH_FILE_NAME". The program is set via a parameters file which is as an input parameter to a command - its path and name has to be specified. The input parameter into the DPE containing the path and name of the parameter file can be omitted if the file is in the same directory as the DPE and its name is ''ParametersFile.txt''. | If your path or name of the parameters includes white spaces enclose the in quotes: "PARAMETERS_FILE_PATH_FILE_NAME". The program is set via a parameters file which is as an input parameter to a command - its path and name has to be specified. The input parameter into the DPE containing the path and name of the parameter file can be omitted if the file is in the same directory as the DPE and its name is ''ParametersFile.txt''. | ||
Revision as of 12:19, 22 June 2023
Data processing enegine serves for a processing of data acquired with the Timepix detectors (TPX, TPX2, TPX3). The following list includes the main processing parts of the DPE:
- Pre-processing: clustering of data with application of calibration and corrections, calculation of cluster variables/parameters, creation of histograms
- Processing: particle recognition
- Post-processing: calculation of physical products - fluxes, dose rates etc.
This document describes how the program can be installed, run, configured etc.
Readme valid for version 1.0.5.
Prerequisites, Install and Run
Program can be dowloaded from this link In the directory, where it should be "installed", download and extract all files in OS directories (Linux or Windows). The ParametersFile.txt includes settings of the program/engine, see description in the next section. There are several other configuration files which sets other processing parts of DPE (histograms, filters, masking etc.) and they are also described in individual section of this document.
To create graphics, python3 and several packages are needed:
- matplotlib (version >= 3.4)
- numpy
- multiprocessing - only if multiprocessing is wanted and CPU has more that one thread
The program especially in the case of graphic export needs several hundreds of MB on RAM (app 500 MB but it is based on the size of exported histograms). If the multiprocessing is used then the RAM usage can be up several GB and all threads of the CPU are used.
Run the program on linux:
chmod +x dpe.sh
./dpe.sh PARAMETERS_FILE_PATH/PARATERS_FILE_NAME
It is possible that also clusterer has to be allowed as an executable: chmod +x clusterer. The current build for linux was made with gcc version 7.5.0 on Ubuntu 18.
Run the program on windows:
DPE.exe PARAMETERS_FILE_PATH\PARATERS_FILE_NAME
If your path or name of the parameters includes white spaces enclose the in quotes: "PARAMETERS_FILE_PATH_FILE_NAME". The program is set via a parameters file which is as an input parameter to a command - its path and name has to be specified. The input parameter into the DPE containing the path and name of the parameter file can be omitted if the file is in the same directory as the DPE and its name is ParametersFile.txt.
First Example of Run
This sections includes information about the first run of the DPE based on the ParamteresFile given with the program. Example directory can be found and downloaded on the google disk which includes needed inputs for the example run on linux and windows systems. After downloading the directories, in the case of the example a combination for linux system: Linux, TPX3Clusterer, Example, the example can be run with following command from the Linux directory:
chmod +x dpe.sh
./dpe.sh ../Example/ParametersFile_Linux.txt
The output should be exported into the Example/Output/Linux directory. The given data is a sample of alpha particles measured with TPX3 Si 500 um detector. Run of the DPE should read the main parameters file ParametersFile_Linux.txt and create several export directories with files and graphics:
- ClusterSensorPlots - individual cluster plots, integrated sensor plots and plots of clusters for individual PID classes.
- Files - main files with the so-called sampling list.
- Graphs - graphical representation of the sampling list.
- Histograms - histograms created based on the Hist.ini configuration file.
- SpatialMaps - spatial maps of cluster variables also with respect of the PID classes.
- SigVec - significant vector which uniquely specifies the given radiation field.
The main configuration is done in the ParametersFile_Linux.txt and meaning of individual parameters can be found there or in this document. The other files in the Example directory determine the creation of histograms (Hist.ini), masking of the raw data (Mask.txt), filtering of cluster (Filter.ini) and creation of the SigVec (SigVec.ini). More information about their format can be found in the next sections in this README.
The similar example can be also made on Windows but with different command from the directory Windows:
DPE.exe ..\Example\ParametersFile_Windows.txt
The output and its description is the same as the for the linux system above.
Additional Examples
Additional examples can be found in the directory Test which includes 10 basic cases of use. The input data are in directory data and its output can be found in the directory out. The examples comprehends following cases:
- Basic t3pa processing from TPX3 detector
- Multi file processing of t3pa files form TPX3 detector
- Clog data from TPX3 measured in tot+toa mode
- Clog data from TPX measured in tot mode
- ClOg data from TPX3 measured in itot+count mode
- Usage of configuration file for histograms Hist.ini on t3pa data
- Usage of configuration file for filtering Filter.ini on t3pa data
- Usage of configuration file for significant vector SigVec.ini on t3pa data
- Usage of configuration file for mask Mask.txt on t3pa data
- Demonstration of distance comparator for radiation filed recognition measured with TPX3 CdTe 2mm
- Elist as input with default settings
- Pregenerated t3pa for physical variables testing with no calibration.
- Combination of SigVec and Hist settings with T3PA file as input to produce SigVec from 2D hist.
All examples were done and produced on linux system but they should be also usable for windows systems.
Issues and Help
Any ideas or bugs can be reported on github forum for issues: https://github.com/lmareksla/DPE_Issues/labels or send directly on the first mail below. A template for the issue can be found on the forum.
For more information or help: lukas.marek@advacam.com, carlos.granja@advacam.com.
Main Configuration of DPE
The main settings of the DPE is done through the so-called parameters file which is a document with all paths, names and switches to configure the DPE. It has similar notation as the c++ code and example can be found in the directory with the program.
There are several other features of DPE which are handled with different configuration files e.g. filtering, histograms etc. Their settings is mentioned in the next sections.
Syntax of the parameters file:
- Everything which is after
//is taken as a comment and the program ignores it (gives opportunity to create comments or easily test different settings . - Every string should be enclosed in
"to correctly account for white spaces in the string. Example:FileName = "file name.txt". - Number can be written in the common notations, e.g. 2e5, 50, -666.12, .1 (same as 0.1) . Example:
Number = 1e7. - If an array of values is needed then it is separated with comma symbol:
,. Example:ArrayNumbers = 12,13, 14, 15,ArrayString = "hop", "skok" - The syntax is partially based on the c++ so it can be used in some highlighting text editors for better clarity.
- All used paths are checked for proper slashes of paths which means that same configuration files can be used on windows and linux. This is not valid for the input argument into DPE = path and name of the configuration file.
List of all adjustable parameters:
- FileIn_Name - [STRING] Name of the input file. Spaces can be part of the name. Example:
FileIn_Name = "tot toa.t3pa". Default: empty string. - FileIn_Path - [STRING] Path of the input file. Example:
FileIn_Path = "/Path/ To/ File/". Default: empty string. - FileIn_NameEnd - [STRING] Name end/extension of the input file. If the file name is
FileIn.txtthen name end is.txt. It is used for a several file processing. Example:FileIn_NameEnd = ".txt". Default: empty string. - FileIn_Count - [INT] Count of input files which should be processed if multi file processing is used. Example
FileIn_Count = 10to process only 10 of input files. Default:-1(unused). - DoMultiFile - [BOOL] Check whether the multi file processing should be used (can be turned off and on). Example:
DoMultiFile = true. Default:false. - CalMatrices_Path - [STRING] Path to a directory with calibration matrices if an input data is without energy calibration. The path should NOT include last
\\(just path to the directory and its name). Example:CalMatrices_Path = "/Path/To/Cal/Matrices". Default: empty string. - SamplingList_Name - [STRING] Name of the sampling list file for export. Example:
FileOutSamplingList_Name = "SamplingList.txt". Default:SamplingList.txt. - FileOut_Path - [STRING] Path of the output file. Example:
FileOut_Path = "/Path/File /Out/". Default: current working directory. - SamplingListJSON_Path - [STRING] Path to output JSON file. Example:
SamplingListJSON_Path = "/Path/File JSON/Out/". Default:SamplingList.json. - SamplingListJSON_Name - [STRING] Name to output JSON file. Example:
SamplingListJSON_Name = "SampligList.json". Default: current working directory. - FileFilter_Name - [STRING] Name of the file with filter configuration. Example:
FileFilter_Name = "Filter.ini". Default: empty string. - FileFilter_Path - [STRING] Path of the file with filter configuration. Example:
FileFilter_Path = "/Path/To/Filter/File/". Default: empty string. - FileMask_Name - [STRING] Name of the file with a mask configuration. Example:
FileMask_Name = "Mask.txt". Default: empty string. - FileMask_Path - [STRING] Path to the file with a mask configuration. Example:
FileMask_Path = "/Path/To/File/With/Mask/". Default: empty string. - FileClusterer_Path - [STRING] Path to clusterer program which is part of the downloaded package. Example:
FileClusterer_Path = "/Path/To/Clusterer/ Program/". Default:../Clusterer/(for linux). - DirModel - [STRING] Path to models. Example:
DirModel = "/Path/To/Models/". Default:./Models/(for linux). - ClogOut_Name - [STRING] Name of the cluster log file for export. If it is stated in the parameters file then the clog is created (alias DoCreateClog = true). Example:
ClogOut_Name = "ClusterLog.txt". Default:ClusterLog.clog. - ElistOut_Name - [STRING] Name of the elist file for export if it is created during raw data processing. Example:
ElistOut_Name = "Elist.txt"(default value also). - TimeSampling - [FLOAT] Time in seconds for individual evaluations of the data sample -> the whole data sample and its time of collection is divided into sampling intervals with duration of the TimeSampling. Example:
TimeSampling = 1e-6to have sampling time of 1 microsecond. Default:1. - PIDAlgSwitch - [INT] Switch to change between different PID algorithms. Example:
PIDAlgSwitch = 101to use first mentioned heuristic decision tree. Default:101.All possibilities (101,102,103,201,202):- 101 - 4 class heuristic decision tree, TPX 300 um Si
- 102 - 9 class heuristic decision tree, TPX 300 um Si
- 103 - 16 class heuristic decision tree, TPX3 500 um Si
- 201 - 3 class DNN, TPX 300 um Si 30 V
- 202 - 6 class DNN, TPX 300 um Si 30 V
- 251 - 3 class DNN, TPX3 500 um Si 80 V
- 252 - 6 class DNN, TPX3 500 um Si 80 V
- DataOriginSwitch - [INT] Currently, it has no effect on the engine run. Example:
DataOriginSwitch = 1to specify environment, ground-level cosmic rays. The engine can characterize general mixed radiation field., Default:0(unused). Number specifying the origin of the data (1,2,3,4):- Environment, ground-level cosmic rays
- Laboratory radionuclides
- Accelerator charged particle beams, low-energy
- Mixed-field space radiation
- DoPrintOut - [BOOL] Control for printout in to the terminal. Example:
DoPrintOut = trueto make print out to the terminal. Default:true. - DoLog - [BOOL] Control for log file. Example:
DoLog = trueto create a log file with program run. Default:true. - DoExportText - [BOOL] Control for an export of the output in txt files. Example:
DoExportText = true. Default:true. - DoExportGraphics - [BOOL] Control for an export of graphical output. Example:
DoExportGraphics = true. Default:false. - DoCreateClog - [BOOL] Control for a generation of the clog. Example:
DoCreateClog = trueif the clog should be created. Default:true. - SensMat - [STRING] Sensor material (Si, CdTe, GaAs). Example:
SensMat = "Si"to specify that the sensor material is silicon. Default:Si. - SensThick - [FLOAT] Sensor thickness in micrometers. Example:
SensThick = 500to specify the thickness of the detector as 500 micrometers. Default:300. - SensBias - [FLOAT] Sensor Bias in volts. Example:
SensBias = 100to specify that applied was 100 V. Default:50. - ChipType - [STRING] Type of the chip (TPX, TPX2, TPX3). Example:
ChipType = "TPX3"to specify that TPX3 was used chip. Default:TPX. - SensMat_Density - [FLOAT] Density of the sensor in g/cm3 for temperature at 20 deg. If not set than its values is deduced based on the type of material but only for the lites ones.
- DoTPXHighEnergyCorr - [BOOL] Switch to use TPX high energy correction. Example:
DoTPXHighEnergyCorr = trueto use high energy correction. Default:false. - DoDoseUnitRadiology - [BOOL] Switch to change from the dose rate in uGy/h to Gy/s. Example:
DoDoseUnitRadiology = trueto use Gy/s. Default:false. - DoRemoveElist - [BOOL] Switch to delete elist created by the clusterer but the extended elist not effected by this switch. If set to true, the elist is removed Example:
DoRemoveElist = truewhich will remove the elist. Default:false. - DoRemoveOldElist - [BOOL] Switch to explicitly delete or not an already existing elist from a previous processing. If set to true, the elist is removed and a new one is created. If false, the DPE checks elist existence and if there is a elist, the pre-processing with clusterer is skipped. Example:
DoRemoveOldElist = true. Default:true. The default value is true because this could cause an error in processing if mask is used with false option -> it wont be processed again and the mask will be ignored for elist data. - DoElistExtended - [BOOL] Switch to produce elist with additional columns from processing output: filter pass or not passed (1,0), PID class (e.g. from -1 to 8). It is placed as the last columns of the elist with name
FilterPass, PIDClass. Example:DoElistExtended = trueto produce this elist. Default:true. - DoCreateExportDir - [BOOL] Switch to produce directory structure for the export. If used, 5 directories are created into which individual files and plots are exported. If not used, everything is exported into one directory. Example:
DoCreateExportDir = trueto produce the directory structure. Default:true. - NClustSensPlot - [INT] Count of clusters which will be exported in the integrated sensor plots (also per class). Example:
NClustSensPlot = 100to export in to the plot 100 clusters (this is the default value). Default:100. - NClustIndivPlot - [INT] Count of clusters which will be exported as individual cluster plots. Example:
NClustIndivPlot = 15to export 15 clusters (this is the default value). Default:15. - NumRebinPlot_Hist1D - [INT] rebin value for 1D histograms before plotting. It will rebin all 1D histograms to make them more clear and exporting graphics faster if value around 1e2 is used. If value 0 is set then this option is skipped. This option overwrite a possible user settings in the configuration files for histograms. Example:
NumRebinPlot_Hist1D = 100all histograms bins will be merge together to show in the graphics only 100 bins (more detailed explanation below). Default:0(no rebinning). - MeasMode - [STRING] Measurement mode of detector = itot+count (itot+count). In the current version, it is used for itot+count mode recognition. Example:
MeasMode = "itot+count"to inform that meas in itot+count was done. Default: empty string. - DoGraphicsMultiProc - [BOOL] Switch to use multiprocessing/treading during graphics export. Example:
DoGraphicsMultiProc = true. Default:true. - GraphicsMultiProc_FileSizeLimit - [FLOAT] File size limit in B on single multi processing batch. It should be decreased if the RAM is overwhelmed during processing. Example:
GraphicsMultiProc_FileSizeLimit = 20e6to set 20 MB. Default:3e6(3 MB).
Example of the ParametersFile can be found in the main directory.
Input
The current version of the DPE engine is capable to process following files:
- Cluster log files- calibrated/uncalibrated
- data driven files - t3pa, t3p, t3r
- Elist - only so-called full elist which includes also the header with cluster variables name.
- Frame formats
If a clog from itot+count measurement is used then it is needed to specify this in the main config file with following option: MeasMode = "itot+count"
Multi file/batch processing
It is allowed to process several raw files. To do that it is needed to specify what is the files first part of the name and ending of the files.
Lets assume a several files: * File_001.t3pa * File_002.t3pa * File_003.t3pa * File.t3pa The DPE has following settings: * FileIn_Name = "File_" * FileIn_NameEnd = ".t3pa" The DPE will only process files which include File_ and .t3pa which is File_001.t3pa, File_002.t3pa and File_003.t3pa but not the last one: File.t3pa. If the FileIn_Name = "File" then also the last file is included. It can be optionally turn off and on with following option: DoMultiFile = true or false. This option is automatically on if the FileIn_NameEnd is non empty from user.
Special settings can be used to find all files ending with given suffix: * FileIn_Name = "" * FileIn_NameEnd = ".t3pa" will find only those files which end with .t3pa.
It is also allowed to processes files in a directory without specifying the name of files with parameters FileIn_Name and FileIn_NameEnd.
In this case, the DPE finds all files with supported suffixes/name end and process those files with the same suffix which are found first.
The only needed parameter is FileIn_Path which leads to a directory with files.
Supported suffixes are following:
".t3pa", ".t3p", ".t3r", ".txt", ".pmf", ".plog", ".bmf", ".clog", ".elist"
Output
During a run of the program, there is an output in to the terminal if the DoPrintOut = true. This include some basic information about the DPE run and its settings. Additionally, during and in the end of processing several text and possibly graphic exports are created.
Text output
- Elist - Document with cluster variables.
- Extended Elist - Elist extended with additional columns of PID class (-1 to N-1 of classes where -1 is for others,
PIDClass) and filter pass (1 = passed, 0 = not passed,FilterPass) if the filter is used. It can be exported with optionDoElistExtended = true. - Cluster log/clog - Detailed list of clusters. Each line which is starting with
[includes pixels of a cluster ([X,Y,E,T] = [X position,Y position,energy,time if tpx3 is sued] of a pixel). This file includes all clusters and the mask or filters are not accounted for. It is reprocessed after every processing of the given file. - Sampling list in txt - Exported to the
FileOut_Pathwith nameFileOutSamplingList_Name. It includes basic information about the radiation field composition with respect to the observables and their time dependencies. The individual recognized components correspond to original settings. - Sampling list in JSON - The same as the txt but in JSON formatting.
- Results of comparator - Results from radiation field recognition with distance comparator (only valid for 2mm CdTe TPX3 data).
- Histograms - Histograms of given variables - total and also for each PID class. More details in a section below.
- Raw data after masking - The DPE can export the original raw data after application of the mask therefore pixels which do not satisfy the mask are removed. This will exported if the mask is used (this is the actual file which is used for data processing, creation of Elist etc.).
- Significant vector - Vector used for field decomposition and other techniques. More details in a section below.
- Cluster and sensor plots - Data files with matrices of exported clusters and sensor.
- Spatial maps - Matrix of the sensor filled with integrated cluster variables (energy, size, LET, E/S).
Graphical output
The current version uses a python scripts to convert the text output into graphics (mainly histograms and graphs of fluxes etc.):
- Histograms
- Spatial maps
- Individual cluster plots (with values of the cluster variables) and integrated sensor plots
- Graphs of time evolution of physical products
For this purposes, the matplolib library is used. It can be slow for some configuration (for example high number of bins). To speed up the export of graphics, it is possible to set shown number of bins with following option: NumRebinPlot_Hist1D = 100 which will cause that each histogram will be rebinned to show maximally 100 bins but the exported text files are still with the original number of bins. This option is only functional for 1D histograms.
Masking
There is a possibility to mask a part of the sensor or individual pixels in the case of t3pa and clog files. These pixels are omitted in the pre-processing.
This can be used to avoid a signal from noisy pixels or to focus on some more interesting part of the sensor.
The configuration is done through a configuration file. An example can be found in the program directory.
Inclusion of Mask.ini into Parametrs File
The configuration file of the mask can be included with following options in the ParametersFile:
- FileMask_Name - Name of the INI configuration file of the mask.
- FileMask_Path - Path of the INI configuration file of the mask.
Syntax of the configuration file:
- The pixel which should be masked is written in format [X,Y]. These are X and Y coordinates of the pixel where both of them are integers from 0 to 255. The sensor orientation is usual and X coordinate is for rows and Y is for lines. They can be written in lines or rows in the mask file but always the [X,Y] in one line (can not be separated over several lines)
- If a larger part is of interest, following notation is used: [X_min - X_max, Y_min - Y_max] where X_min is the minimum coordinates of a pixel, X_max is the maximum coordinate of a pixel etc. For example, [0-255,0-120] masks almost half of the sensor - on the X axis from 0 to 255 and on the Y axis from 0 to 120. A simpler demonstration can be masking of one line of pixels (11th) : [0-255, 10].
Example of all possibilities for masking:
[0,1] [2,5] [2,3] [2-43,5] [76,8-90] [0-50,50-60]
If the masking is used the during the pre-processing an additional t3pa/clog file is created with masked pixels according to the user configuration in the export directory (starts with MASK_+ FileIn_Name). The value of masking should be larger than 255.
Filtering
During the processing, filters can be used to obtain only information about particles of interest. The filters are applied on cluster variables/parameters level (e.g. energy, height etc.). It is specified trough a configuration file in the INI format. An example can be found in the program directory. The cluster variables which should be used for filtering are specified with their unique name which is included in the header of the created elist (if elist is input then it has to be already part of the file).
Inclusion of Filter.ini into Parametrs File
The configuration file of the filter can be included with following options in the ParametersFile:
- FileFilter_Name - Name of the INI configuration file of the filter.
- FileFilter_Path - Path of the INI configuration file of the filter.
Syntax of the configuration INI file:
- If the energy filter should be used then in the file is place
[E]alias key name of the filter. - Individual conditions are then written as
Range_1=100,200. This settings will produce a filter on cluster energy which should be only from 100 to 200 keV (edges are included). - More ranges can be specified for one variable. See example below:
Range_1=100,200
Range_2=500,1000
Range_asdasd=300,2000
- The range values and name of the parameters should always include string
Range. If more ranges are used then their parameter names should differ but always include stringRange, for exampleRange_1,Range_2,Range_N.
The list all possible cluster variables which can be used for ranges is following:
#define NAME_DETECTOR_ID "DetectorID" //1 - detector ID
#define NAME_EVENT_ID "EventID" //2 - event ID (coincidence ID)
#define NAME_FLAGS "Flags" //3 - user flag
#define NAME_X "x" //4 - X weighted center
#define NAME_Y "y" //5 - Y weighted center
#define NAME_TIME_MIN "t" //6 - min time
#define NAME_SIZE "Size" //7 - size
#define NAME_ENERGY "E" //8 - deposited energy
#define NAME_HEIGHT "Height" //9 - energy of pixel with maximum energy
#define NAME_BORDER_PIX_N "BorderPixCount" //10 - count of border pixels
#define NAME_ROUND "Roundness" //11 - roundness
#define NAME_LIN "Linearity" //12 - linearity
#define NAME_POLAR_ANGLE "Angle" //13 - polar angle
#define NAME_L2D "Length" //14 - projected length
#define NAME_WIDTH "Width" //15 - projected width
#define NAME_LET "LET" //16 - linear energy transfer
#define NAME_EPIX_MEAN "EpixMean" //17 - averaged energy per pixel
The last two are not default part of the elist but they are calculated during the processing.
Histograms
One of the DPE outputs are also histograms of cluster variables. It is possible to export 2D and 1D histograms which can be configured with a configuration file in the INI format (an example can be found in the program directory).
The DPE allows to also create histograms of algebraic combinations of the cluster variables (multiplication, division, subtraction, addition).
It is important that the general label (key name of a section in a INI file) of the histogram used in the INI file is unique to each histogram. If there are more histograms with one common name then the program only updates information about the first one in the INI file. The label itself in not used in the program itself and title and name of the histogram are set separately.
Inclusion of Hist.ini into Parameters File
The the DPE will include a user Hist.ini file only if it is written in the PararamtersFile. Two parameters are used for this purpose:
- FileHist_Name - Name of the configuration file for histograms.
- FileHist_Path - Path of the configuration file for histograms.
If both of them are set and the file is found on the given location then the DPE uses this settings of histograms otherwise default configuration is used.
Histogram 1D:
The 1D histograms can be created as fixed bin width histograms or with variable binning. The fixed bin width has following settings/needed parameters (example from configuration file with explanations = everything after #):
VarName="Size" # Name of column with given variable (see the first line in Elist.txt - it has to be the same)
Title="S" # Title of histogram (can be arbitrary - X if not given)
NBin=9 # Number of bins
Xmin=100 # Minimum value (if X = Xmin -> it is NOT included to the first bin - it has to be > Xmin)
Xmax=1000 # Maximum value (if X = Xmax -> it is included to last bin NBin)
The same as above but with different choice of variable, it is based on the column position in the elist instead of the cluster variable name:
ColIndex=7 # Position of the column - starts from 0 (it is Size in the example)
Title="S" # -||-
NBin=9 # -||-
Xmin=100 # -||-
Xmax=1000 # -||-
These cases create histogram from cluster size with 9 bins from 100 to 1000 where one bin has width 100. Variable size bin width and its needed parameters (same histogram as the one above):
VarName="Size" # -||-
Title="S" # -||-
BinLowEdge=100,200,300,400,500,600,700,800,900,1000 # Low edges of bins with Xmax (Xmin,...,Xmax -> size NBin+1)
Histogram 2D:
The 2D histograms are constructed in similar manner as 1D histograms. The fixed bin width histograms and their needed parameters:
VarName="E","Size" # Name of columns with given variable - 1st is X, 2nd is Y (see the first line in Elist.txt - it has to be the same or see special variables)
Title="E,S" # Title of histogram (can be arbitrary - X if not given)
NBinX=9 # Number of X bins where X is in this case energy,E
Xmin=100 # Minimum value of X (if X = Xmin -> it is NOT included to first bin - it has to be > Xmin)
Xmax=1000 # Maximum value of X (if X = Xmax -> it is included to last bin NBin)
NBinY=1000 # Number of Y bins
Ymin=0 # Minimum value of X (if X = Xmin -> it is NOT included to first bin - it has to be > Xmin)
Ymax=1000 # Maximum value of X (if X = Xmax -> it is included to last bin NBin)
An example for cluster variables based on column positions in the elist:
ColIndex=4,7 # Position of the columns which should be processed - 1st is X, 2nd is Y
...(the same as above)
These cases create 2D histogram of cluster energy and size where energy is from 100 to 1000 with bin width of 100 and size is from 0 to 1000 with bin width of 1. Variable size bin width and its needed parameters:
VarName="E","Size" # -||-
Title="E,S" # -||-
BinLowEdgeX=100,200,300,400,500,600,700,800,900,1000 # Low edges of X bins with Xmax (Xmin,...,Xmax -> size NBinX+1)
BinLowEdgeY=100,200,300,400,500,600,700,800,900,1000 # Low edges of Y bins with Ymax (Ymin,...,Ymax -> size NBinY+1)
It is also possible do just variable binning in one variable the second one can be with fixed bin size:
VarName="E","Size"
Title="E,S"
NBinX=9
Xmin=100
Xmax=1000
BinLowEdgeY=100,200,300,400,500,600,700,800,900,1000
Additional algebraic operations:
Both 1D and 2D histograms allow additional operations of addition, subtraction, multiplication and division on extracted variables from EList.
Histogram 1D:
ColIndex_Div=4,7 # Division as 5th/7th column (in this case E/S = Epix) - 1st argument is divided by 2nd argument
ColIndex_Mult=4,7 # Multiplication as 5th*7th column - 1st argument is multiplied with 2nd argument
ColIndex_Add=4,7 # Additions 5th+7th column- 1st argument is added to 2nd argument
ColIndex_Subtr=4,7 # Subtraction as 5th-7th column - 2nd argument is subtracted from 1st argument
An example based on cluster variable names:
VarName_Div="E","Size" # Same thing as above but with names of columns
VarName_Mult="E","Size" # Same thing as above but with names of columns
VarName_Add="E","Size" # Same thing as above but with names of columns
VarName_Subtr="E","Size"# Same thing as above but with names of columns
Histogram 2D:
Very similar to 1D histograms but always the first given is X and the second given is Y.
ColIndex=4 # X is set to 4th column - energy
ColIndex_Div=4,7 # Y is set to division of 4th/7th
Operations used for both variables:
ColIndex_Div=8,7,4,7 # X is set to division of 8th/7th and Y is set to division of 4th/7th
The same thing can be done with names of columns/variables VarName:
VarName_Div="E","Size","Height","Size" # X is E/Size and Y is Height/Size
Text export
There are two kinds of exported files: histogram data and histogram info file. The histogram data file includes content of bins and bins low edges. The histogram info file comprehends features of the histogram: title, name, count of bins etc. (see more details below). The data file has a suffix .hist and the info file .hist_info. The data file has following formatting for 1D histogram:
Xmin BinCont_1
BinLowEdge_2 BinCont_2
... ....
BinLowEdge_NBin BinCont_NBin
Xmax Overflow
The Xmax is included to allow an user easier read of this file without direct need to also read the info file (all needed information is in the data file in the base case). It is also possible to export only those bins which have non zero content with fixed binning. This can be done with following option in the Hist.ini file:
- DoSparseExport - Check whether only nonzero bins should be exported (1 for true and 0 for false).
It has to be used for each histogram separately and it is used as default settings. The exported data file has following format:
Xmin BinCont_1
BinLowEdge_2 BinCont_2
... ...
BinLowEdge_i BinCont_i != 0
... ...
Xmax Overflow
The Xmin,BinLowEdge_2,Xmax are always exported even if their bin content is 0 for further reading and reconstruction of histograms. This option is not functional for variable binning to avoid a lack of information in the data file for histogram complete reconstruction in post-processing. To reconstruct the histogram, it can be done just based on the data file even in the case of sparse export because the missing bins and bin width can be calculated based on the first two bins in the data file (BinWidth = BinLowEdge_2 - Xmin). Anyway, it is recommended to read the info file for example in the case of constant bin width combined with the sparse export to ensure that given histogram is truly with constant binning and not variable binning (written in the parameter: BinEquiDist=1 = it is const binning and 0 for variable binning).
Similar approach is utilized for the 2D histograms:
Xmin Ymin BinCont_1 # First bin
XBinLowEdge_2 Ymin BinCont_2
... .... ....
XBinLowEdge_NBinX Ymin BinCont_NBinX
Xmin YBinLowEdge_2 BinCont_NBinX+1
... .... ....
XBinLowEdge_NBinX YBinLowEdge_2 BinCont_NBinX+NBinY
XBinLowEdge_2 YBinLowEdge_3 BinCont_NBinX+NBinY+1
... .... ....
XBinLowEdge_NBinX YBinLowEdge_NBinY BinCont_NBinX*NBinY # Last bin
Xmax Ymax Overflow
The sparse export has following form.
Xmin Ymin BinCont_1
XBinLowEdge_2 Ymin BinCont_2
... .... ....
XBinLowEdge_i Ymin BinCont_i != 0
... .... ....
Xmin YBinLowEdge_2 BinCont_NBinX+1
... .... ....
XBinLowEdge_m YBinLowEdge_n BinCont_m+n*NBinX != 0
... .... ....
Xmax Ymax Overflow
It is similar to 1D histogram with the exception that there is also the next Y low bin edge to also estimate the width of binning for Y axis.
The info file is formatted as an INI file and it includes the same information as the Hist.ini file for each histogram individually. There two additional features compared with the Hist.ini:
- Statistical information (mean, err of mean, std, err of std)
- Overflow and underflow information
Graphical export
There is possibility to create plot of Hist1D and Hist2D via python matplotlib, it has to be preinstalled. The current version will automatically create these plots if DoExportGraphics=true but it can be turned off - see below.
SOME_PREVIOUS_CODE # This is some code to call histogram 2D
Name="Histogram2D title in plot" # Name o histogram in plot
AxisTitle="X","Y","N" # Name of axis in the plot
The plots can be with logarithmic scales on all axis:
SOME_PREVIOUS_CODE # This is some code to call histogram 2D
DoLogX=1 # 1 for true=do logarithmic X axis and 0 for false
DoLogY=1 # 1 for true=do logarithmic Y axis and 0 for false
To turn off plotting:
SOME_PREVIOUS_CODE # This is some code to call histogram 2D
DoPlot=0 # Default is 1 - do plot and 0 means not do plot
It is possible to adjust the number binning of the histogram only for the graphics. This can be done with NumRebinPlot_Hist1D in the main configuration file:
NumRebinPlot_Hist1D = 100
This option with value 100 will change the shown number of bins in the graphics to 100 but the exported files includes original binning. This is allowed for faster exports of histograms with more than 10000 bins which can time challenging for a PC.
Significant Vectors
The significant vectors is a unique set/vector of numbers which describes given data sample/radiation field. It can be used for radiation field recognition as it is done in the DPE. There is a possibility to create own configuration file for the generation of the significant vector.
Inclusion of SigVec.ini into Parameters File
The the DPE will include a user configuration file SigVec.ini only if it is written in the PararamtersFile. Two parameters are used for this purpose:
- FileSigVec_Name - Name of the configuration file for significant vectors.
- FileSigVec_Path - Path of the configuration file for significant vectors.
If at least the name is set and the file is found on the given location then the DPE uses this settings of SigVec otherwise default configuration is used.
Histogram 1D - Intervals of Interest
A histogram is examined and all bins (bin contents) with bin center between up and down edge/limit of given interval is summed, number R_1. This number is used as one element of SigVec = {R_1, R_2, ... , R_N} for N of intervals. At least two numbers has to be given - down and up edge of the interval where interval condition for bins are following: DOWN_EDGE < BinCenter <= UP_EDGE. An example of configuration file for 1D histogram:
[Var_Epix] #Section name as unique key of the given SigVec element. It has to include the string "Var_".
Title="Epix" #Title of the variable.
HistName="Hist_Epix" #Name of histogram which should be processed - same as name if histogram file without end (data end)
Intervals=0,200,100,500,500,5000 #Limits of intervals - 1st interval = from 0 to 200, 2nd interval = from 100 to 500 etc. (N in this case is 3)
The intervals can be overlapping. The section name has to "Var" include string in its name and they have to differ for different elements of the SigVec: Var_E, Var_S, ....
Histogram 2D - Regions of Interest
A histogram is multiplied with mask (from 0 val to inf - can serve as weights) with the same number of bins. Then the histogram is summed and this number is as an input to the SigVec. For each given mask one number is produced. An example of configuration file:
[Var_E_S] #Section name as unique key of the given SigVec element. It has to include the string "Var_".
Title="E_S" #Title of the variables
HistName="Hist_E_S_2" #Name of histogram which should be processed - same as name if histogram file without suffix (data end = .hist)
Mask_1="\PATH\TO\MASK\MASK1_FILE_NAME" #Path to masks which should be applied - same dimensions as histogram (see examples)
Mask_2="\PATH\TO\MASK\MASK2_FILE_NAME" #Path to masks which should be applied - same dimensions as histogram (see examples)
Lets assume that the Hist_E_S_2 is following histogram: NBinX = 5, Xmin = 0, Xmax = 10 & NBinY = 4, Xmin = 0, Xmax = 4 then the mask should have following form:
0 1 0 0 0 0 0 0 0 0 0 2 0 0 3 0 0 0 4 10
A single gap is used as number separator.This will use bins [X,Y,Weight] = [2,1,1], [2,3,2], [5,2,3], [4,4,4], [4,5,10] and the matrix is read from first line to the last one (1st line is Y = 1).
Normalization
Both 1D and 2D histograms can be normalized before they are used calculation of the SigVec:
Normalize=1 #1 to DO normalization - 0 to NOT DO norm. - default is 0
Particle Identification
The DPE engine is also capable of basic particle identification.
This output can be exported into extended elist whose last column/PIDClass will include the information about recognized class.
frame
The choice of the PID algorithm is based on the switch PIDAlgSwitch whose numerical value are listed below. The settings can be done in the main configuration file:
PIDAlgSwitch = 101
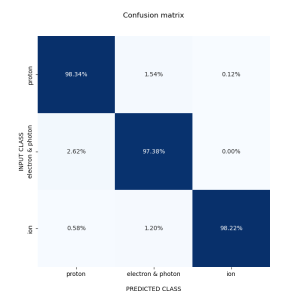
the current possible values are following: * 101 - Heuristic simplified DT for TPX 300 um Si * 102 - Heuristic decision tree with 8 classes for TPX 300 um Si * 103 - Heuristic decision tree with 16 classes for TPX3 500 um Si * 201 - Dense neural network with 3 classes for TPX 300 um Si * 202 - Dense neural network with 6 classes for TPX 300 um Si * 251 - Dense neural network with 3 classes for TPX3 500 um Si * 252 - Dense neural network with 6 classes for TPX3 500 um Si If not value is given to the program then DT is chosen based on the detector configuration.
Dense neural networks for TPX 300 um Si
- Dense neural network TPX with 3 classes
- Protons
- Photons && Electrons
- Ions
- Others
- Dense neural network TPX with 6 classes
- Protons LE (<30 MeV)
- Protons ME (>30 & <100 MeV)
- Protons HE (>100 MeV)
- Photons && Electrons
- Helium ions
- Ions (except He)
- Others
Dense neural networks for TPX3 500 um Si
- Dense neural network TPX3 with 3 classes
- Protons
- Photons && Electrons
- Ions
- Others
- Dense neural network TPX3 with 6 classes
- Protons LE (<30 MeV)
- Protons ME (>30 & <100 MeV)
- Protons HE (>100 MeV)
- Photons && Electrons
- Helium ions
- Ions (except He)
- Others
Decision tree for TPX 300 um Si
- Heuristic simplified DT with 3 classes:
- Low LET: electrons, X-rays, gamma
- Mid LET: protons
- High LET: ions
- Others
- Heuristic decision tree with 8 classes:
- X-rays; LE electrons OD; HE electrons OD; muons PP
- LE protons OD; HE protons PP
- LE alphas OD; HE alphas PP
- LE ions OD; HE ions PP
- HE electrons OD; muons nPP
- HE protons nPP; UHE protons nPP; UHE alphas nPP
- He alphas nPP; UHE light ions nPP
- He ions nPP
- Others
Decision tree for TPX3 500 um Si
- Heuristic decision tree with 16 classes:
- X-rays; HE electrons PP; HE protons PP
- Gamma rays LE (e.g. 137 Cs)
- Gamma rays HE (e.g. 60 Co)
- LE electrons OD (< 10 MeV)
- HE electrons nPP (> 10 MeV)
- LE and HE electrons PP
- LE protons (< 3MeV)
- ME protons (3-10 MeV)
- HE protons (>10 MeV)
- LE alphas (<10 MeV)
- HE alphas (>10 MeV)
- LE ions (<10 MeV/u)
- HE ions (>10 MeV/u)
- LE, thermal and slow neutrons ( < 0.5 eV)
- HE fast neutrons ( > 1 MeV)
- Others
Explanation of used abbreviations: LE - Low Energy, HE - High Energy, UHE - Ultra High Energy ,OD - Omni Directional, PP - Perpendicular to the sensor, nPP - non Perpendicular. Their values differentiate for individual decision tree.
Radiation Field Recognition
The DPE is capable of basic radiation filed recognition (RFR). Several algorithms can be used for this purposes:
- Distance Comparator
To use the RFR, it is needed to use standard/default settings of the DPE with respect to the histograms and significant vectors.
Distance Comparator
The distance comparator uses the significant vectors of premesured known radiation fields (database of vectors) and compare them with the given data for processing. There has to be match between the database detector configuration and the data configuration. The comparison is based on distance evaluation between the known and unknown vector. The final probability that given data is one of the source is based on inproportional relation between probability and the distance. You can find below currently included databases for the detector configuration.
CdTe 2mm TPX3 and efficiency results:
IN/OUT Ba133 Cs137 Eu152 Co60 Am241 Na22
Ba133 97.40 0.40 1.31 0.26 0.33 0.30
Cs137 3.22 77.73 4.51 4.56 2.40 7.57
Eu152 2.70 1.08 94.16 0.62 0.69 0.74
Co60 1.06 2.53 1.29 89.48 1.22 4.41
Am241 1.47 1.37 1.56 1.31 93.04 1.24
Na22 0.87 3.13 1.10 3.28 0.81 90.81
Post-processing and Physical Products
The main physical products which can be found in the exported files are following:
- Flux
- Count of particles and pixels
- Dose rate
- Deposited energy
- Distributions of cluster variables
- Spatial event maps of cluster variables
- Spatial map of clusters and individual clusters
The flux and dose rate account for possible masking. In the output sampling list, all time coupled variables are in the cases of classes calculated with respect to the sampling time. The total variables are calculated with respect to the elapsed time which is usually shorter. The elapsed time is defined as time of the last processed event/particle. Therefore these values are bigger than the sum/mean of variables of classes.
Sampling of data
Most of the physical products are calculated based on a sampling time. This means that all events/particles which are within a time interval of sampling time from some starting point contribute to the physical products.
Example, lets assume that measurement of 10 s were done and 20000 particles were registered in first 5 seconds and 30000 particles in another 5 seconds. If the sampling time is chosen as 5 s than 2 samples are created in the output file with two values for each sampled physical product. The values of flux, if no mask is used, are then: 2000 and 3000 particles/s cm-2. The total flux is calculated based on the elapsed time and if the last event was detected at time of 9 s then the total flux is: (20000 + 30000 particles)/(2 cm2 * 9 s) = 2777,8 particles/s cm-2 which is more than the mean value of the class fluxes 2500 particles/s cm-2.
Detector Settings
One of the DPE parameters allow to set the detector configuration:
- SensMat - sensor material (Si, CdTe, GaAs etc). Example:
SensMat = "Si"to specify that the sensor material is silicon. Possible values:Si- for silicon sensor. Used density:CdTe- for cadmium teluride sensor. Used density:GaAs- for galium arsenide. Used density:
- SensThick - Sensor thickness in micrometers. Example:
SensThick = 500to specify the thickness of the detector as 500 micrometers. This is continues variable. - SensBias - sensor bias in volts. Example:
SensBias = 100to specify that applied was 100 V. This is a continues variable. - ChipType - type of the chip (TPX or TPX3). Example:
ChipType = "TPX3"to specify that TPX3 was used chip. Possible values:TPX- for timepix chipTPX3- for timepix 3 chipTPX2- for timepix 2 chip
These values are used, for example, in the calculation of dose rate because there is a dependence on the sensor material and thickness. It also determines which PID algorithm should be used.
Error Codes and Others
Error Codes
From version 1.0.6.
The engine run can produce an error code into standard error stream (stderr) and into the standard output stream (stdout).
Successful engine run produces/returns 0. Any other value signals error in the run.
Possible values are listed below (negative numbers with explanation after //):
#define DPE_ERR_NO_ERROR 0 //No error occurred. #define DPE_ERR_NO_PAR_FILE -1000 //Missing file with parameters. #define DPE_ERR_INIT_GEN -1001 //Initialization has not been done. #define DPE_ERR_READ_PAR_GEN -1002 //UNUSED #define DPE_ERR_OPEN_LOG -1003 //Can not open log file. #define DPE_ERR_NO_IN_FILE -1004 //Can not find any files for processing. #define DPE_ERR_NO_IN_FILE_SEL_IGN -1005 //No files have passed the selection and ignore criteria. #define DPE_ERR_IN_FILE_TYPE -1006 //File can not be opened or unknown format (binary). #define DPE_ERR_IN_SIMPLE_ELIST -1007 //Input data recognized as ElistSimple. Can not be processed because the names of variables are missing. #define DPE_ERR_NO_INIT_PID -1008 //PID init has not been done. #define DPE_ERR_FIND_CLUSTERER -1009 //Can not find clusterer binary. #define DPE_ERR_FIND_MODELS -1010 //Can not find models directory. #define DPE_ERR_CHECK_PYTHON -1011 //Missing python for graphics creation. #define DPE_ERR_CHECK_PYTHON_MOD -1012 //Missing python modules for graphics creation (names are in stderr and stdout). #define DPE_ERR_NO_MASK_FILE -2000 //File with mask can not be opened. #define DPE_ERR_LOAD_CLOG_TACQ -2100 //Can not load acq time of frames from input clog. #define DPE_ERR_NO_ELIST -3000 //Elist file can not be opened. #define DPE_ERR_NO_EELIST -3001 //ExtElist can not be opened. #define DPE_ERR_NO_T_PREV -3002 //Missing previous time in elist processing. #define DPE_ERR_EXP_NO_DATA -4000 //No data was processed. Can not continue with export. #define DPE_ERR_OPEN_SLIST -4001 //Can not open sampling list. #define DPE_ERR_OPEN_SLIST_ALL -4002 //Can not open sampling list for overall info. #define DPE_ERR_OPEN_SLIST_FL -4003 //Can not open sampling list for flux info. #define DPE_ERR_OPEN_SLIST_ED -4004 //Can not open sampling list for Edep info. #define DPE_ERR_OPEN_SLIST_DR -4005 //Can not open sampling list for DR info. #define DPE_ERR_OPEN_SLIST_JSON -4006 //Can not open sampling list in json. #define DPE_ERR_OPEN_SLIST_JSON_ALL -4007 //Can not open sampling list overall in json. #define DPE_ERR_OPEN_SLIST_JSON_FL -4008 //Can not open sampling list flux in json. #define DPE_ERR_OPEN_SLIST_JSON_ED -4009 //Can not open sampling list Edep in json. #define DPE_ERR_OPEN_SLIST_JSON_DR -4010 //Can not open sampling list DR in json. #define DPE_ERR_RELOAD_CLOG -4011 //Can not open file with clog for clusters and sensor plots. #define DPE_ERR_EMPTY_CL -4012 //Cluster list is empty for clusters and sensor plots. #define DPE_ERR_GRAPH_MULTIP_CDIR -4013 //Exporting graphics was not successful in the multiprocessing part (open curr dir). #define DPE_ERR_GRAPH_MULTIP_0FUNC -4014 //Exporting graphics was not successful in the multiprocessing part (no sub function for processing). #define DPE_ERR_GRAPH_MULTIP_RFUNC -4015 //Exporting graphics was not successful in the multiprocessing part (can not read sub function file). #define DPE_ERR_RELOAD_CLOG_OPENF -4016 //Can not open file with clog for clusters and sensor plots. #define DPE_ERR_RELOAD_CLOG_NUPIX -4017 //Incorrect number of pixel data in line with cluster. #define DPE_ERR_CLUSTMATRIX_EXPCL -4018 //Can not export maximal coincidence group because the storage is empty. #define DPE_ERR_DEL_ELIST -4023 //Can not delete elist file.